api¶
MRS.api¶
Functions and classes for representation and analysis of MRS data. This is the main module to use when performing routine analysis of MRS data.
GABA¶
- class MRS.api.GABA(in_data, w_idx=[1, 2, 3], line_broadening=5, zerofill=100, filt_method=None, spect_method={'BW': 2, 'NFFT': 1024, 'n_overlap': 1023}, min_ppm=-0.7, max_ppm=4.3, sampling_rate=5000.0)¶
Bases: object
Class for analysis of GABA MRS.
Methods
- __init__(in_data, w_idx=[1, 2, 3], line_broadening=5, zerofill=100, filt_method=None, spect_method={'BW': 2, 'NFFT': 1024, 'n_overlap': 1023}, min_ppm=-0.7, max_ppm=4.3, sampling_rate=5000.0)¶
Parameters: in_data : str
Path to a nifti file containing MRS data.
w_idx : list (optional)
The indices to the non-water-suppressed transients. Per default we take the 2nd-4th transients. We dump the first one, because it seems to be quite different than the rest of them.
line_broadening : float (optional)
How much to broaden the spectral line-widths (Hz). Default: 5
zerofill : int (optional)
How many zeros to add to the spectrum for additional spectral resolution. Default: 100
filt_method : dict (optional)
How/whether to filter the data. Default: None (#nofilter)
spect_method: dict (optional) :
How to derive spectra. Per default, a simple Fourier transform will be derived from apodized time-series, but other methods can also be used (see nitime documentation for details)
min_ppm, max_ppm : float
The limits of the spectra that are represented
sampling_rate : float
The sampling rate in Hz.
- est_gaba_conc()¶
Estimate gaba concentration based on equation adapted from Sanacora 1999, p1045
Ref: Sanacora, G., Mason, G. F., Rothman, D. L., Behar, K. L., Hyder, F., Petroff, O. A., ... & Krystal, J. H. (1999). Reduced cortical
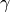 -aminobutyric acid levels in depressed patients determined by
proton magnetic resonance spectroscopy. Archives of general psychiatry,
56(11), 1043.
-aminobutyric acid levels in depressed patients determined by
proton magnetic resonance spectroscopy. Archives of general psychiatry,
56(11), 1043.
- fit_creatine(reject_outliers=3.0, fit_lb=2.7, fit_ub=3.5)¶
Fit a model to the portion of the summed spectra containing the creatine and choline signals.
Parameters: reject_outliers : float or bool
If set to a float, this is the z score threshold for rejection (on any of the parameters). If set to False, no outlier rejection
fit_lb, fit_ub : float
What part of the spectrum (in ppm) contains the creatine peak. Default (2.7, 3.5)
- fit_gaba(reject_outliers=3.0, fit_lb=2.8, fit_ub=3.4, phase_correct=True, fit_func=None)¶
Fit either a single Gaussian, or a two-Gaussian to the GABA 3 PPM peak.
Parameters: reject_outliers : float
Z-score criterion for rejection of outliers, based on their model parameter
fit_lb, fit_ub : float
Frequency bounds (in ppm) for the region of the spectrum to be fit.
phase_correct : bool
Where to perform zero-order phase correction based on the fit of the creatine peaks in the sum spectra
fit_func : None or callable (default None).
If this is set to False, an automatic selection will take place, choosing between a two-Gaussian and a single Gaussian, based on a split-half cross-validation procedure. Otherwise, the requested callable function will be fit. Needs to conform to the conventions of fit_gaussian/fit_two_gaussian and ut.gaussian/ut.two_gaussian.
- fit_glx(reject_outliers=3.0, fit_lb=3.6, fit_ub=3.9, fit_func=None)¶
Fit a Gaussian function to the Glu/Gln (GLX) peak at 3.75ppm, +/- 0.15ppm [Hurd2004]. Compare this model to a model that treats the Glx signal as two gaussian peaks. Glx signal at. Select between them based on cross-validation
Parameters: reject_outliers : float or bool
If set to a float, this is the z score threshold for rejection (on any of the parameters). If set to False, no outlier rejection
fit_lb, fit_ub : float
What part of the spectrum (in ppm) contains the GLX peak. Default (3.5, 4.5)
scalefit : boolean
If this is set to true, attempt is made to tighten the fit to the peak with a second round of fitting where the fitted curve is fit with a scale factor. (default false)
References
[Hurd2004] (1, 2) 2004, Measurement of brain glutamate using TE-averaged PRESS at 3T
- fit_glx2(reject_outliers=3.0, fit_lb=3.6, fit_ub=3.9, phase_correct=True, scalefit=False)¶
Parameters: reject_outliers : float or bool
If set to a float, this is the z score threshold for rejection (on any of the parameters). If set to False, no outlier rejection
fit_lb, fit_ub : float
What part of the spectrum (in ppm) contains the creatine peak. Default (3.5, 4.2)
scalefit : boolean
If this is set to true, attempt is made to tighten the fit to the peak with a second round of fitting where the fitted curve is fit with a scale factor. (default false)
- fit_naa(reject_outliers=3.0, fit_lb=1.8, fit_ub=2.4, phase_correct=True)¶
Fit a Lorentzian function to the NAA peak at ~ 2 ppm. Example of fitting inverted peak: Foerster et al. 2013, An imbalance between excitatory and inhibitory neurotransmitters in amyothrophic lateral sclerosis revealed by use of 3T proton MRS
- fit_water(line_broadening=5, zerofill=100, filt_method=None, min_ppm=-5.0, max_ppm=5.0)¶
- reset_fits()¶
This is used to restore the original state of the fits.
- voxel_seg(segfile, MRSfile)¶
add voxel segmentation info
Parameters: segfile : str
Path to nifti file with segmentation info (e.g. XXXX_aseg.nii.gz)
MRSfile : str
Path to MRS nifti file
SingleVoxel¶
- class MRS.api.SingleVoxel(in_file, line_broadening=5, zerofill=100, filt_method=None, min_ppm=-0.7, max_ppm=4.3)¶
Bases: object
Class for representation and analysis of single voxel (SV) -PROBE experiments.
- __init__(in_file, line_broadening=5, zerofill=100, filt_method=None, min_ppm=-0.7, max_ppm=4.3)¶
Parameters: in_file : str
Path to a nifti file with SV-PROBE MRS data.
line_broadening : float
How much to broaden the spectral line-widths (Hz)
zerofill : int
How many zeros to add to the spectrum for additional spectral resolution
min_ppm, max_ppm : float
The limits of the spectra that are represented
fit_lb, fit_ub : float
The limits for the part of the spectrum for which we fit the creatine and GABA peaks.